Problems of high dimension
- Many ML algorithm relies on distances to compute similarity between samples
- Curse of dimensionality – as number of dimension increases, distances between any two sample from the dataset tends to be more alike (more concentrated) in a way that makes it hard to differentiate them. Every point is point is roughly in similar distance to each other, which results in more difficulty in grouping/cluster the samples
- Risk of overfitting – the model might try to fit to the irrelevant features (noise) just because of some noise in the training set but such relationship is not present in the validation/test data set
- Can even add more noise during prediction phase for testing
- Resource waste – more features means more columns of data to store/process/maintain
- Performance – the extra features can cause more time to process for an online system
- Redundant features can cost problem of collinearity
- which is harmful to some model like linear regression
- it’s hard for the model to be confident in the coefficients of two high correlated features, which results in high p-value of both coefficients
- which is harmful to some model like linear regression
- Features space grows exponentially for each feature added, which requires a lot data to train
- Feature space become sparse
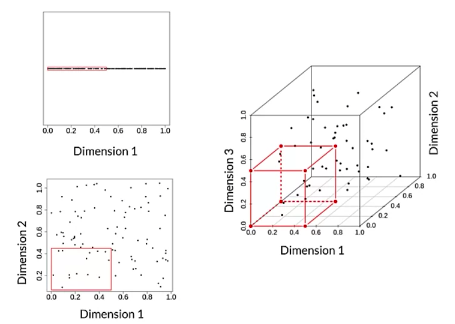
- The Hughes effect – as number of dimension increases, it’s harder to find correct hypothesis (because the hypothesis space is larger) and it needs more examples to generalize
Algorithms
- Latent Semantic Indexing/Analysis (LSI/LSA)
- Unsupervised
- Single Value Decomposition (SVD)
- For decomposing non-square matrix A = S*V*D
- Reconstruct original matrix with few dimensions
- Principle Component Analysis (PCA)
- Only for square matrices
- Eigen value decomposition
- Only for square matrices
- Independent Component Analysis (ICA – paper)
- Separates multivariate signal into addictive components that are maximally independent
- Used for separating super-imposed signals
- Difference with PCA
- PCA – looks for uncorrelated factors
- ICA – looks for independent factors (a factor is uncorrelated to any other factors)
- Not result in orthogonal components like PCA
- Removes high order dependence
- Non-negative Matrix Factorization (NMF)
- Sample features need to be non-negative
- Latent Dirichlet Allocation (LDA)
Other dimension reduction technique
- Quantization
- This is a technique rather than a specific algorithm
- Benefit
- Lower cpu/gpu/memory/power consumption
- Faster inference performance/latency
- Made possible to run on mobile/embedded devices
- Disadvantage
- Might lose precision
- Values might be saturated/clipped if the quantization range is not set up correctly
- Quantize the input
- Ex: reduce a color image to a gray scale or even black and white image.
- Quantize model weight
- Use lower precision model weight and operations (float -> 8 bit)
- Input support is more available in embedded devices
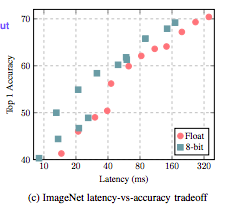 from: Quantization and Training of Neural Networks for Efficient Integer-Arithmetic-Only Inference
from: Quantization and Training of Neural Networks for Efficient Integer-Arithmetic-Only Inference- Type of model quantization
- Quantize model post training (support in Tensorflow Lite)
- Very simple to apply
- Quantization aware training (in Tensorflow)
- Need to add fake quantization nodes during training (see two “quant” nodes below)
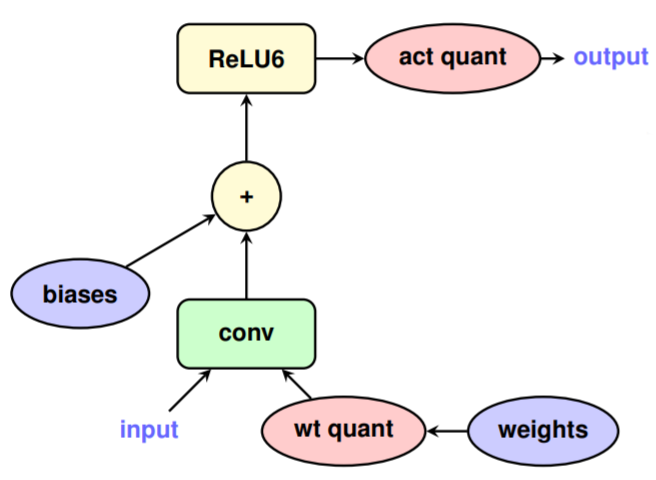
- Quantize model post training (support in Tensorflow Lite)
- Pruning
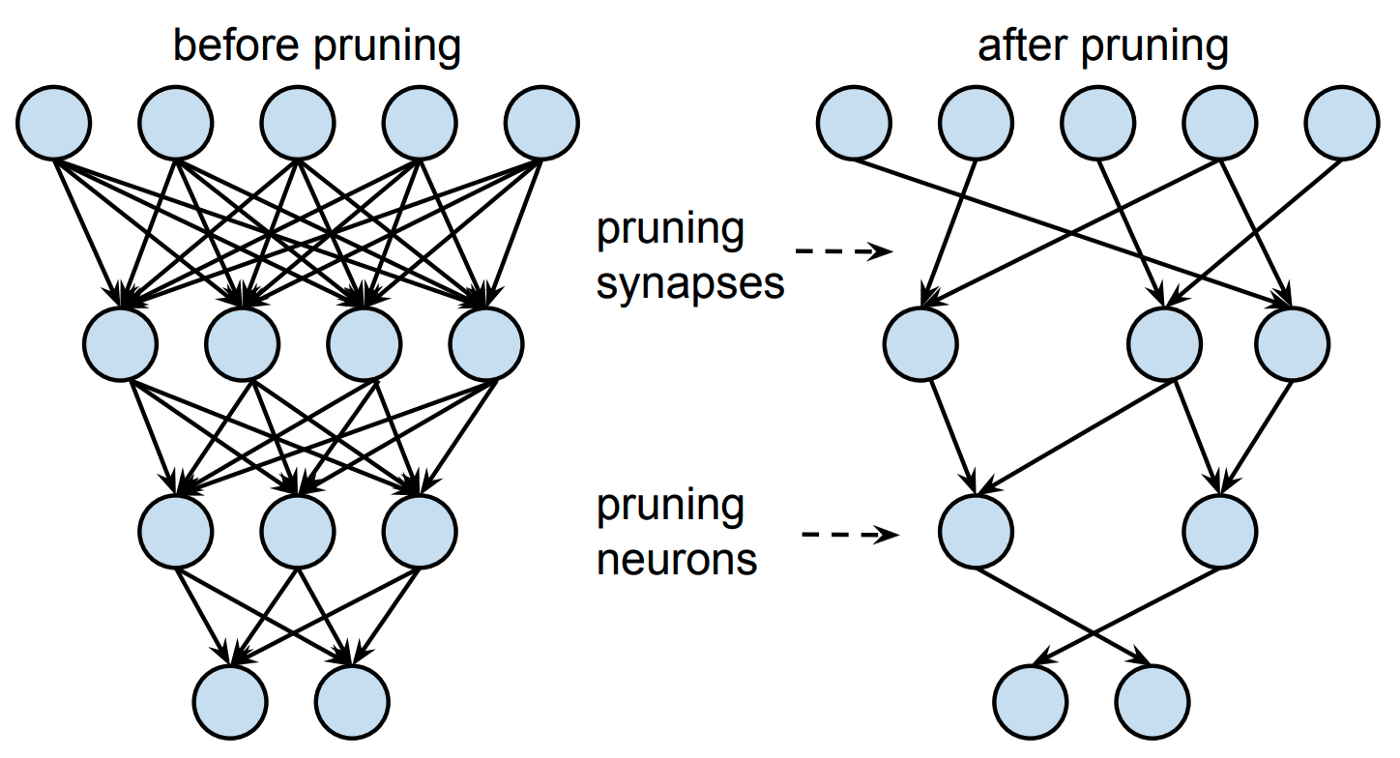
- Cutting out nodes/edges in a neural network.
- Reduces number of parameters (small model to store)
- Reduces number of edges/connections (fewer operation to process)
- Act as a form of regularization
- Weight pruning based on magnitude during training
- Slow ramp up pruning using a schedule until desired sparsity is reached
- Lottery ticket hypothesis
- An over-parameterized network contains a subnetwork that perform as well as the entire network
- Observation: Post pruning + fine tuning might worse than just using original weights
- There is a subnetwork that “won the lottery”
- Sparse tensor in tensorflow
The unavoidable
Sometimes you just have to use a large model to squeeze every bit of performance. You might need to use distributed training when the model is too large to fit in a machine (Training large models – distributed training)
One thought on “Dimensionality Reduction in Machine Learning”